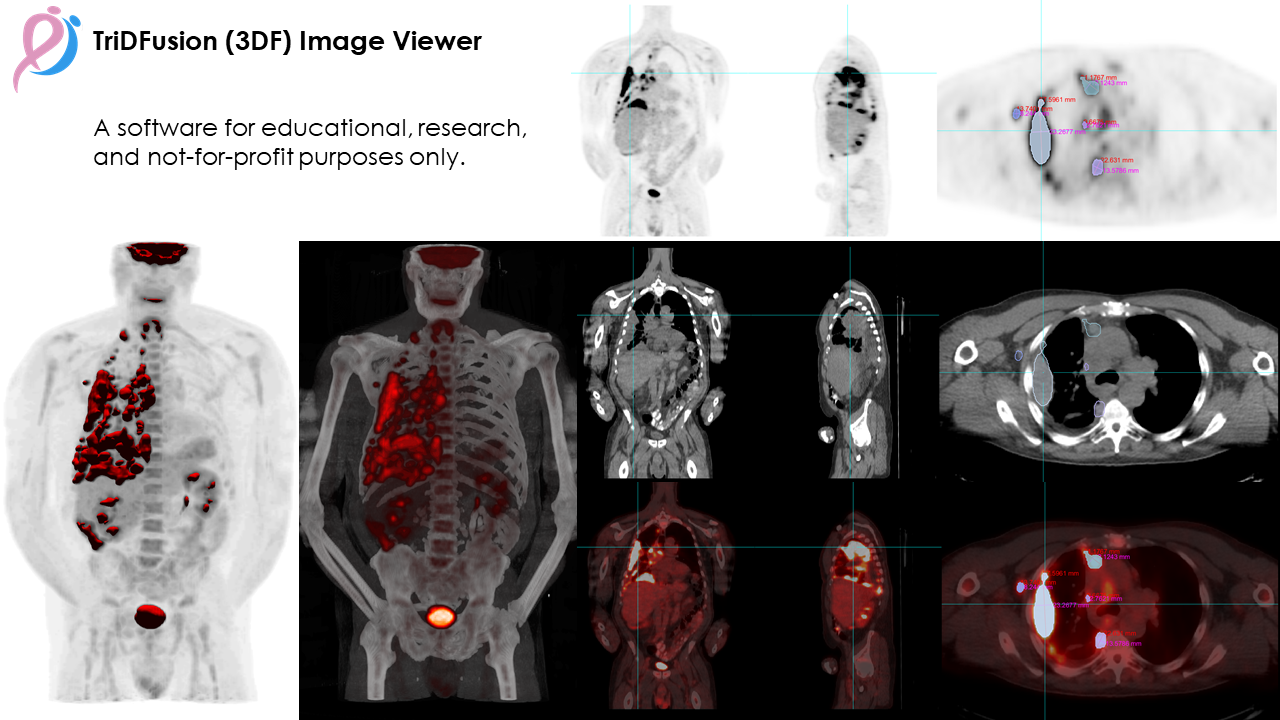
The TriDFusion (3DF) Image Viewer is an advanced, feature-rich research tool built specifically for physicists and medical imaging researchers, offering comprehensive support for various medical imaging modalities. Designed to handle multi-dimensional DICOM images, this powerful viewer integrates dynamic memory allocation to ensure smooth, high-performance processing of large datasets. The 3DF Viewer addresses a range of common imaging challenges, such as orientation errors, slice misalignment, and registration inconsistencies, enabling researchers to focus on precise analysis. Additionally, it includes capabilities for adjusting and removing hot pixels, which is especially beneficial in high-noise images, like Whole-Body SPECT planar bladder scans.
Supported Modalities and File Formats
The 3DF Image Viewer supports a wide variety of medical imaging modalities, including Positron Emission Tomography (PET-CT), Nuclear Medicine (NM), Computed Tomography (CT), Digital Radiography (CR, DX), Digital Angiography (XA), Magnetic Resonance (MR), Mammography (MG), Ultrasonography (US), and others. It also handles import and export across multiple formats, including DICOM (with custom/vendor dictionaries), DICOM-RT structure files, raw nuclear imaging data, and advanced export options like CERR planC dose volumes and constraints. It further supports export to common image formats, such as JPEG, PNG, BMP, and specialized research formats like Neuroimaging Informatics Technology Initiative (NIfTI) and Nearly Raw Raster Data (NRRD), allowing researchers flexibility in data sharing and interoperability.
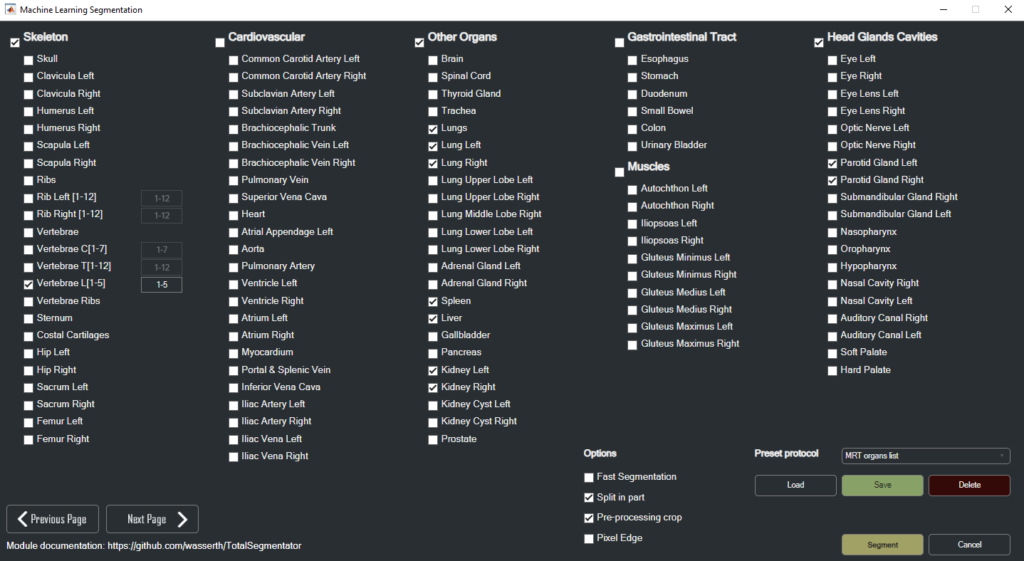
Enhanced Segmentation and Masking Tools
Among its standout features, the 3DF Image Viewer offers sophisticated tools for masking and image segmentation across all modalities, enabling researchers to precisely delineate areas of interest such as tumors, organs, or critical structures. Segmented images can be exported as new DICOM series or in .stl format, enabling applications in 3D printing for physical modeling of complex anatomical structures.
Fusion, Registration, and Visualization
The 3DF Viewer is equipped with multi-modality image fusion tools that allow users to combine MIP with isosurface imaging, improving lesion detection and characterization, particularly in 18F-FDG PET/CT studies. Enhanced volume rendering with customizable color and transparency maps enables researchers to visualize uptake intensity, facilitating deeper insight into pathology. It also includes robust image registration, re-orientation, convolution, and resampling capabilities, allowing seamless alignment and preparation of images for accurate analysis and fusion. For users needing more advanced image manipulations, the viewer supports image arithmetic, post filtering, and edge detection for refined adjustments.
Advanced AI and Machine Learning Toolkits
For tumor quantification, 3DF includes a specialized AI toolkit that allows segmentation results to be directly exported to machine learning frameworks, such as NN-Unet, for use in deep learning applications. This is particularly valuable for those using AI to improve the precision of tumor segmentation and quantification. The toolkit extends to machine learning applications for 3D lung shunt analysis, lung lobe quantification, Y90 dosimetry, and automated lung segmentation, enhancing the software’s utility in both oncology and dosimetry research.
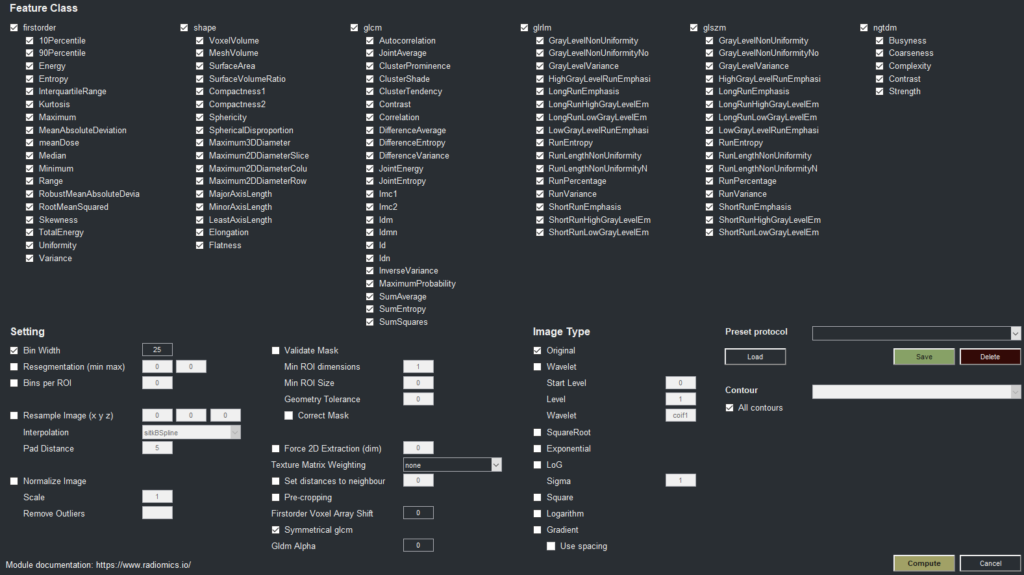
Quantitative Analysis and Radiomics
The 3DF Viewer includes sophisticated tools for total tumor burden determination, a critical factor in cancer research and treatment planning, allowing researchers to accurately quantify and monitor tumor volumes over time. The software’s radiomics capabilities enable researchers to extract a broad array of quantitative imaging features, supporting predictive modeling and personalized treatment approaches in nuclear medicine and radiology.
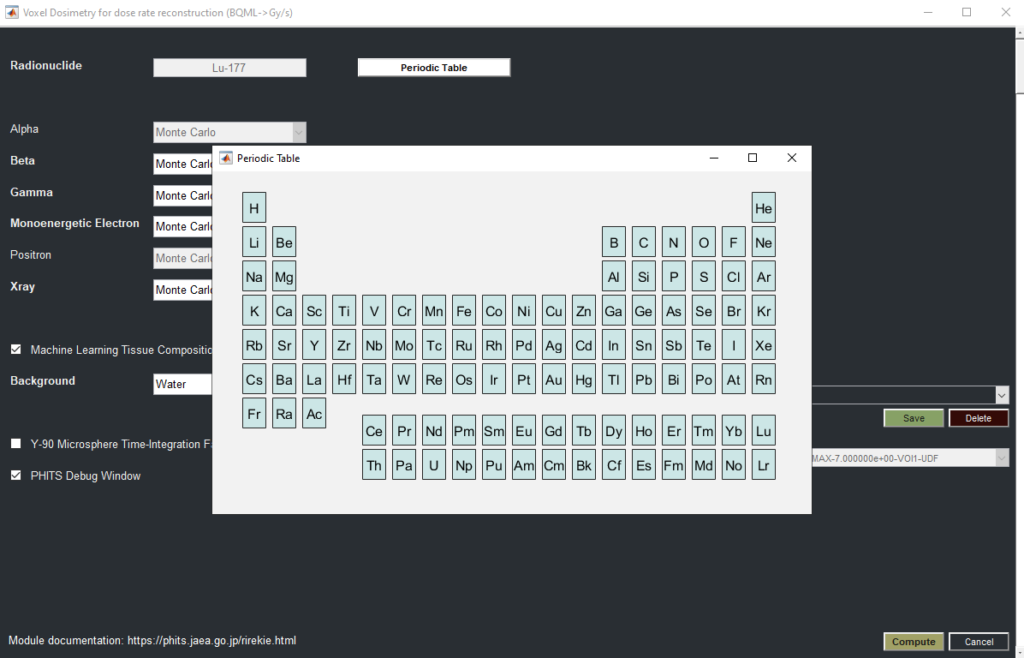
Voxel-Based Dosimetry and Clinical Applications
The viewer also offers voxel-based dosimetry for various isotopes, empowering researchers to calculate precise dose distributions and customize dosimetry calculations for individual patients. In clinical research settings, the 3DF’s tools support image constraints, lung segmentation, and lung dose calculations, enabling more tailored and precise dosimetric analysis.
With its expansive array of features, including 3D visualization, multi-fusion, lung segmentation, image editing, and radiomics, the TriDFusion (3DF) Image Viewer stands as an essential tool for medical physicists, imaging researchers, and academic professionals. By integrating advanced imaging capabilities with AI-driven segmentation, quantitative analysis, and seamless format compatibility, 3DF is revolutionizing workflows and advancing the field of medical imaging research.
The TriDFusion (3DF) image viewer is published in the EJNMMI Physics journal and is accessible online here.
If you use TriDFusion (3DF) please cite it:
Lafontaine D, Schmidtlein CR, Kirov A, Reddy RP, Krebs S, Schöder H, Humm JL. TriDFusion (3DF) image viewer. EJNMMI Phys. 2022 Oct 18;9(1):72. doi: 10.1186/s40658-022-00501-y. PMID: 36258098; PMCID: PMC9579267.
| Medical Imaging Modalities | Import / Export File Formats |
|---|---|
| Positron Emission Tomography PET-CT (PT) | DICOM using custom/vendor dictionaries |
| Gamma Camera, Nuclear Medicine (NM) | Raw data from nuclear imaging devices |
| Computed Tomography (CT) | DICOM-RT structure (contours) |
| Digital Radiography (CR, DX) | CERR planC, dose volumes and constraints |
| Digital Angiography (XA) | Comma Separated Values (.csv) |
| Magnetic Resonance (MR) | Standard Triangle Language (.stl) |
| Secondary Pictures and Scanned Images (SC) | Audio Video Interleave (.avi) |
| Mammography (MG) | Moving Pictures Expert Group 4 (.mp4) |
| Ultrasonography (US) | Graphics Interchange Format (.gif) |
| Digital Imaging and Communications (.dcm) | |
| Joint Photographic Experts Group (.jpg) | |
| Bitmap (.bmp) | |
| Portable Network Graphics (.png) | |
| Neuroimaging Informatics Technology Initiative (.nii) | |
| Nearly Raw Raster Data (.nrrd) |
Main features:
- Multi-modality Image Viewer
- Total Tumor Burden Determination
- 3D Visualization
- 3D Printing
- Image Multi-Fusion
- Image Convolution
- Image Registration
- Image Resampling
- Image Re-Orientation
- Image Arithmetic and Post Filtering
- Image Editing
- Image Mask
- Image Constraint
- Lung Segmentation
- Edge Detection
- Voxel Dosimetry
- Machine Learning Segmentation
- Machine Learning 3D Lung Shunt & Lung Dose
- Machine Learning 3D Lung Lobe Quantification
- Machine Learning Y90 Dosimetry
- Radiomics
Visit GitHub to download the source code.
Matlab compiled version can be downloaded here.
Examples:
Publications:

Lafontaine D, Schmidtlein CR, Kirov A, Reddy RP, Krebs S, Schöder H, Humm JL. TriDFusion (3DF) image viewer. EJNMMI Phys. 2022 Oct 18;9(1):72. doi: 10.1186/s40658-022-00501-y. PMID: 36258098; PMCID: PMC9579267.

Daniel Lafontaine, Charles Schmidtlein, Adam Kesner, Assen Kirov, Joseph O’Donoghue and John Humm
Journal of Nuclear Medicine June 2023, 64 (supplement 1) P495
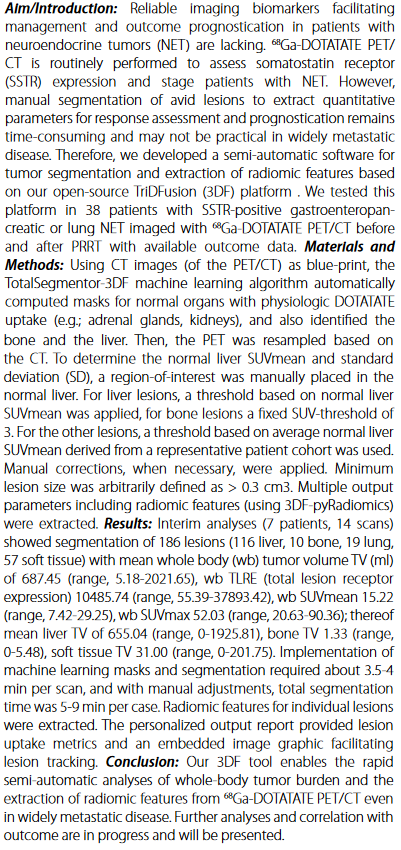
Krebs, Simone; Lafontaine, D; Bodei, L; Mayerhoefer, ME; Humm, J; Schoder, H (2023). Deakin University. Journal contribution. https://hdl.handle.net/10779/DRO/DU:24945177.v1

Daniel Lafontaine, Harry Marquis, Adam Kesner, Assen Kirov, Olga Talarico, Simone Krebs, Constantinos Sofocleous, Heiko Schoder, John Humm and Lukas Carter
Journal of Nuclear Medicine June 2024, 65 (supplement 2) 241493;

Daniel Lafontaine, Finn Augensen, Adam Kesner, Raoul Vincent, Assen Kirov, Simone Krebs, Heiko Schöder & John L. Humm
EJNMMI Phys 12, 22 (2025). https://doi.org/10.1186/s40658-025-00732-9
Project Lead and Developer
Daniel Lafontaine*
Contributing Team
C. Ross Schmidtlein*, Brad Beattie*, Assen Kirov*, Aditya Apte*, Adam Kesner*, Joseph O’Donoghue*, Alexandre Franca Velo*, Simone Krebs**, Lukas Carter*
Memorial Sloan Kettering
*Department of Medical Physics, New York, NY, USA
**Department of Radiology, New York, NY, USA
← Pojects